Germplasm Gernotype
Germplasm Genotype Page
This independent page URL: http://www.mbkbase.org/{species}/genotype/byName/{name}
{species} : species name, such as "rice".
{name}: sample or germplasm name
This page includes three models:
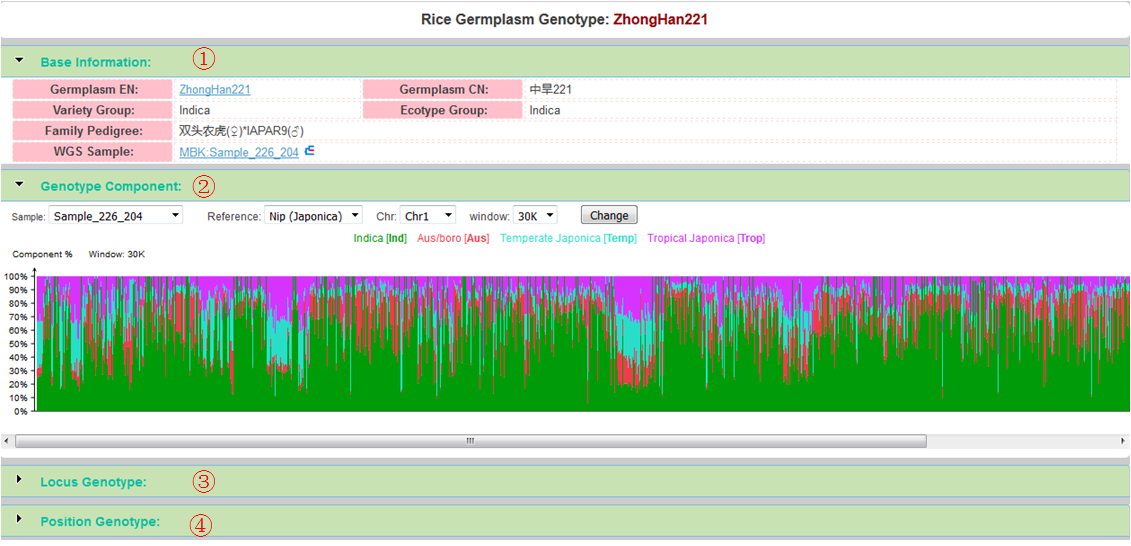
- Base Information (see below)
- Genotype Component
- Locus Genotype
- Position Genotype
Locus Genotype
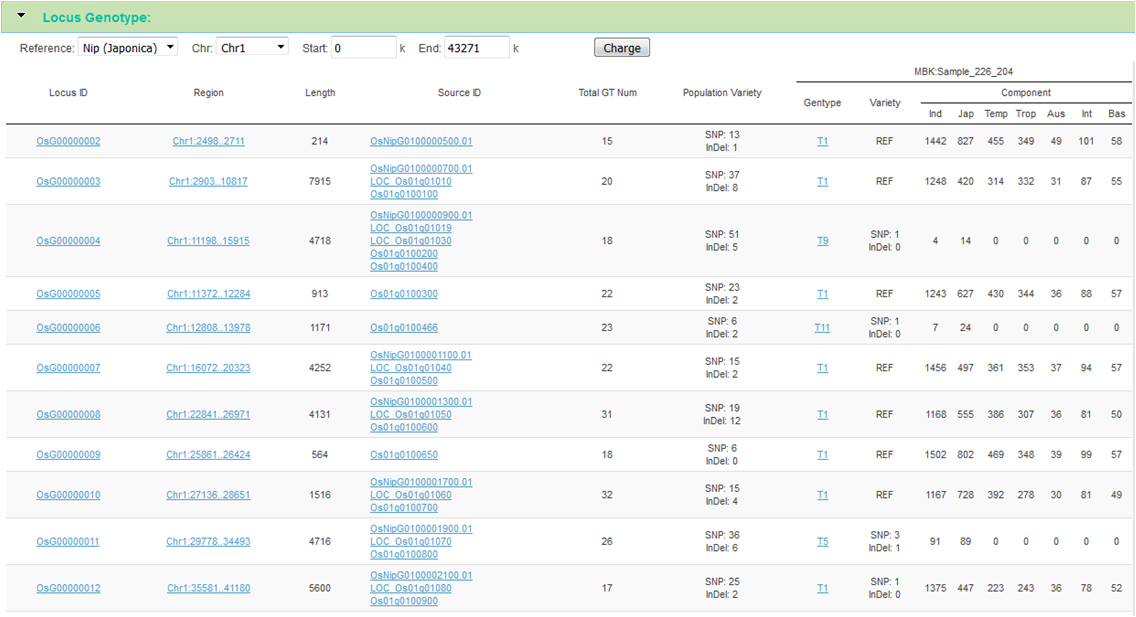
Locus Genotyping and Show:
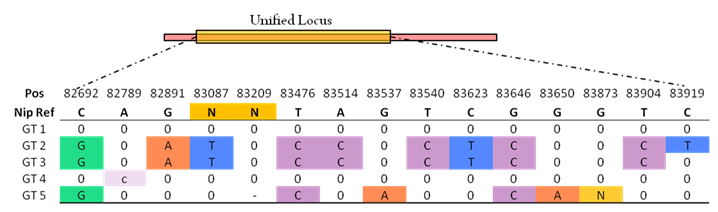
In our rice genotype database, a Locus genotype is an absolute measure of base composition of a group of WGS samples (germplasms) in a unified locus region. One genotype (Locus GT) is same or differs subtly from reference genome sequence, and the differs including both SNP and InDel, but not other big structural variations. Capital base means this position variation is homozygous, lowercase is heterozygous, '-' mean missing reads. Because of most cultivated rice germplasm are homozygous, so the rice genotype in our database is same as the conception of haplotype and allele.For each potentially variant site, both SNP and InDel with allele frequency >0.15% were retained for building genotype. In Ref row, charts 'N' represents multiple continuous bases, which means the variation of this position for corresponding sample is deletion, in genotype row, the 'N' represents insertion of bases.
Position Genotype
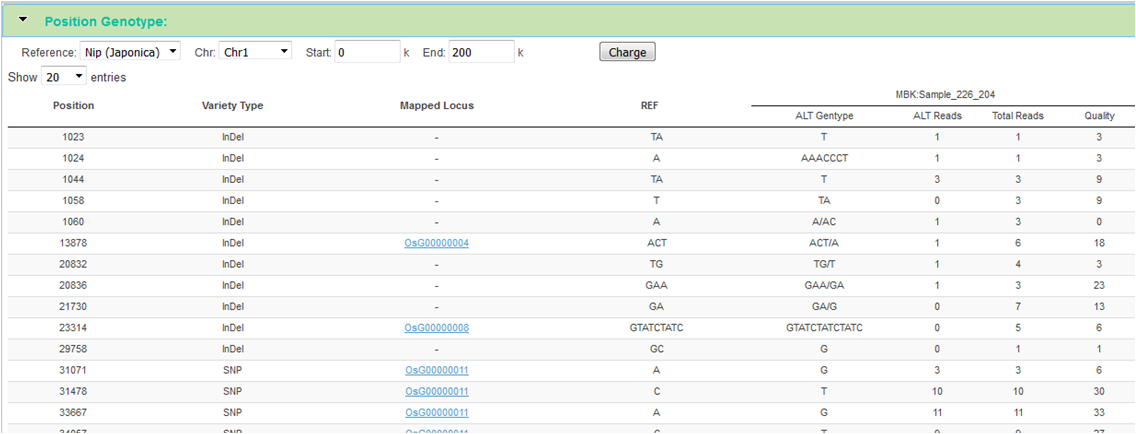
In our database, a position genotype (ALT) is an absolute measure of base composition in a reference genome position. One genotype (Position GT) is a SNP or InDel, the heterozygous mutations using the symbol "/" division, and the homozygous mutations not contain the symbol.
Created with the Personal Edition of HelpNDoc: Single source CHM, PDF, DOC and HTML Help creation